Accession
MI0000821
Symbol
MGI:
Mir133b
Description
Mus musculus
mmu-mir-133b precursor miRNA
Literature search
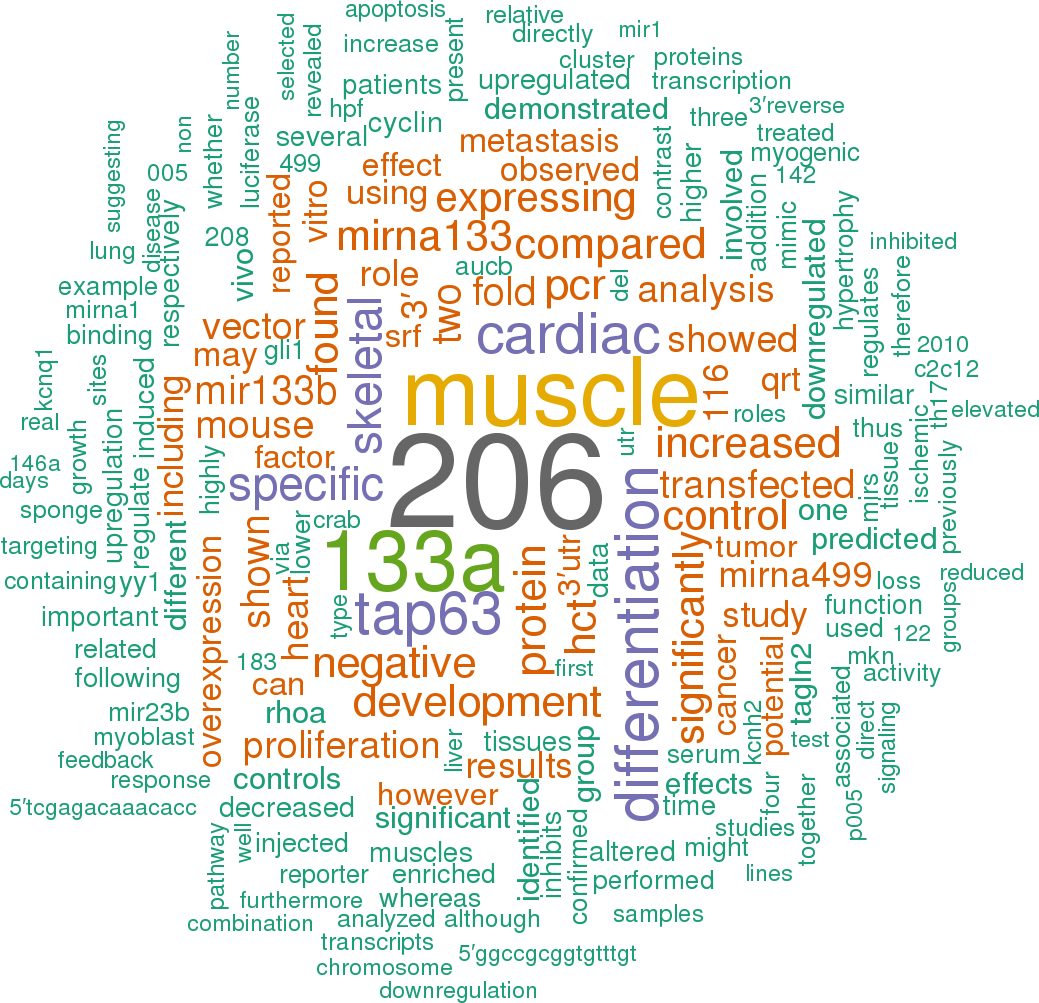
198 open access papers mention mmu-mir-133b
(1312 sentences)
(1312 sentences)
Sequence
118935
reads,
1590
reads per million, 71 experiments
ccuccaaagggaguggcccccugcucugGCUGGUCAAACGGAACCAAGUCcgucuuccugagaggUUUGGUCCCCUUCAACCAGCUAcagcagggcuggcaaagcucaauauuuggaga
.(((((((..((((.((((((((((.((((((((..((.((.((((((.((.((((...)))))))))))).)).))..)))))))).)))))))..)))...))))....))))))).
.(((((((..((((.((((((((((.((((((((..((.((.((((((.((.((((...)))))))))))).)).))..)))))))).)))))))..)))...))))....))))))).
Structure
c --gg --g -- c CA C A U g c cuccaaa gagu gcc cccugcu ugGCUGGU AA GG ACCAAG Cc ucuu ||||||| |||| ||| ||||||| |||||||| || || |||||| || |||| c gagguuu cucg cgg gggacga AUCGACCA UU CC UGGUUU gg agag a auaa aaa uc c AC C C - - u
Annotation confidence
High
Do you think this miRNA is real?
Comments
miR-133b was predicted based on comparative analysis of human, mouse and Fugu [1]. It is homologous to miR-133a (MIR:MI0000159 and MIR:MI0000820). The mature sequence shown here represents the most commonly cloned form from large-scale cloning studies [3]. The 5' end of the miRNA may be offset with respect to previous annotations.
Genome context
chr1: 20682769-20682887 [+]
Clustered miRNAs
1 other miRNA is < 10 kb from mmu-mir-133b
| Name | Accession | Chromosome | Start | End | Strand | Confidence |
|---|
Mature mmu-miR-133b-5p
| Accession | MIMAT0017083 |
| Description | Mus musculus mmu-miR-133b-5p mature miRNA |
| Sequence | 29 - GCUGGUCAAACGGAACCAAGUC - 50 |
| Evidence |
experimental
Illumina [5] |
| Database links |



|
| Predicted targets |


|
Mature mmu-miR-133b-3p
| Accession | MIMAT0000769 |
| Description | Mus musculus mmu-miR-133b-3p mature miRNA |
| Sequence | 66 - UUUGGUCCCCUUCAACCAGCUA - 87 |
| Evidence |
experimental
cloned [2-3], Illumina [4-5] |
| Database links |



|
| Predicted targets |


|
References
|



