Accession
MI0006977
Description
Oryza sativa
osa-MIR444e precursor miRNA
Gene family
MIPF0000402;
MIR444
Literature search
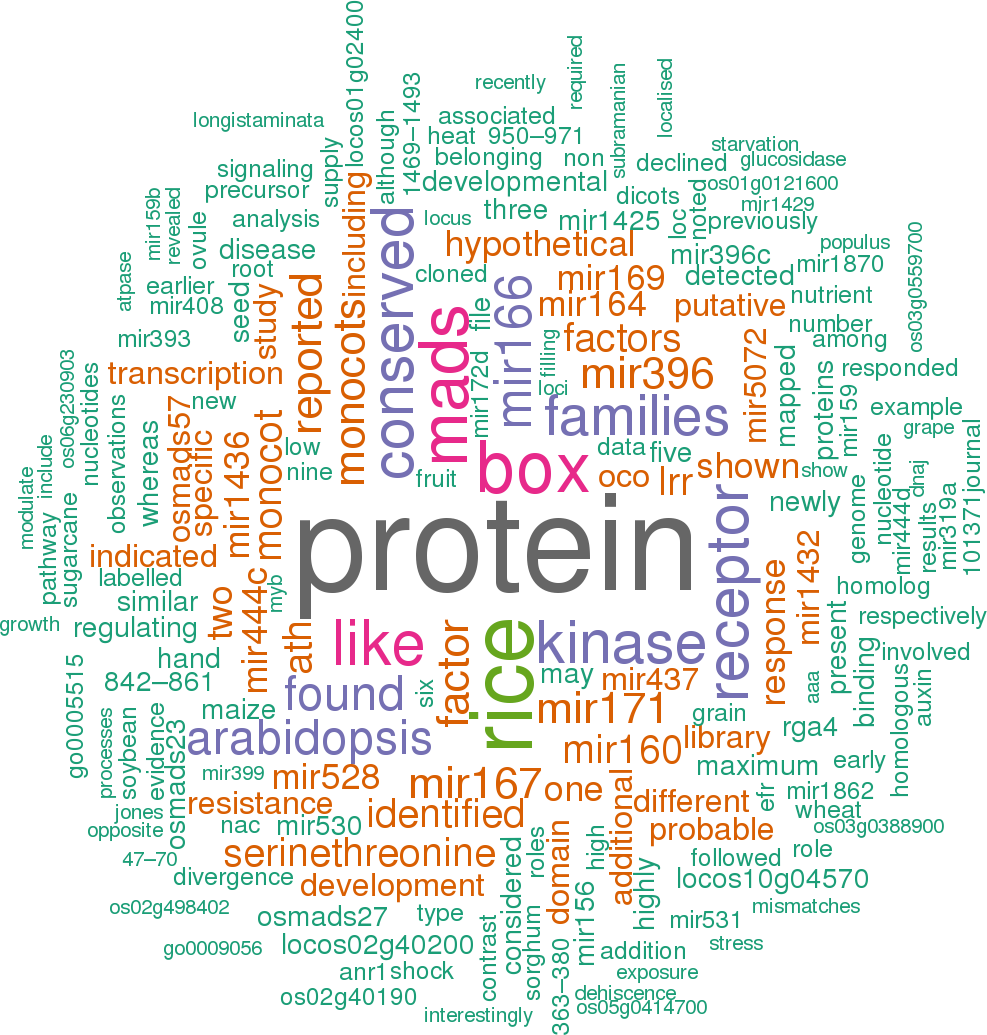
23 open access papers mention osa-MIR444e
(67 sentences)
(67 sentences)
Sequence
2473
reads,
891
reads per million, 2 experiments
gguuuuguaaccaggggcaauaagcuagaggcaccaacugcauaacuugcgagaaacuuguuggaugaugaucuugccaaugauuuugccgcaaguuaUGCAGUUGCUGCCUCAAGCUUacugccuuuguuugcuaaacc
(((((.((((.((((((((.((((((.((((((.(((((((((((((((((..(((.(((((((............))))))).)))..))))))))))))))))).)))))).)))))).)))))))).)))).)))))
(((((.((((.((((((((.((((((.((((((.(((((((((((((((((..(((.(((((((............))))))).)))..))))))))))))))))).)))))).)))))).)))))))).)))).)))))
Structure
u c a a c ag c augau
gguuu guaa caggggca uaagcu gaggca caacugcauaacuugcg aaa uuguugg g
||||| |||| |||||||| |||||| |||||| ||||||||||||||||| ||| |||||||
ccaaa cguu guuuccgu aUUCGA CUCCGU GUUGACGUauugaacgc uuu aguaacc a
u u c A C cg u guucu
Annotation confidence
Not enough data
Do you think this miRNA is real?
Genome context
Unknown
Database links
Mature osa-miR444e
| Accession | MIMAT0005975 |
| Description | Oryza sativa osa-miR444e mature miRNA |
| Sequence | 99 - UGCAGUUGCUGCCUCAAGCUU - 119 |
| Evidence |
experimental
MPSS [1], Northern [1], cloned [2] |
| Database links |

|
References
|

