Accession
MI0000165
Symbol
MGI:
Mir140
Description
Mus musculus
mmu-mir-140 precursor miRNA mir-140
Gene
family?
family?
RF00678;
mir-140
Literature search
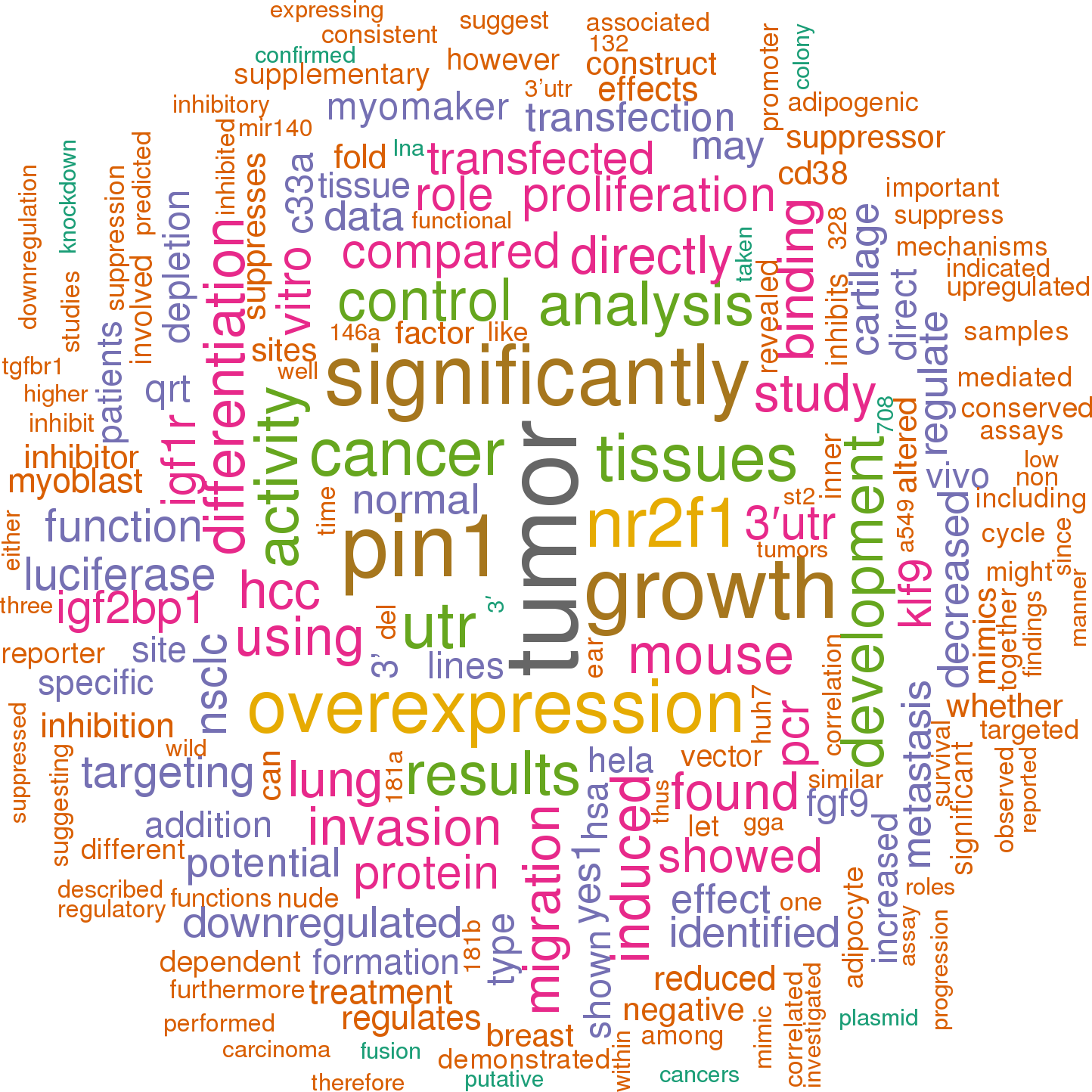
88 open access papers mention mmu-mir-140
(916 sentences)
(916 sentences)
Sequence
7001290
reads,
16032
reads per million, 107 experiments
ccugcCAGUGGUUUUACCCUAUGGUAGguuacgucaugcuguucUACCACAGGGUAGAACCACGGacagg
((((((.(((((((((((((.(((((((.((.((...)))).))))))).))))))))))))))).))))
((((((.(((((((((((((.(((((((.((.((...)))).))))))).))))))))))))))).))))
Structure
- A A u c c
ccug cC GUGGUUUUACCCU UGGUAGg ua gu
|||| || ||||||||||||| ||||||| || || a
ggac GG CACCAAGAUGGGA ACCAUcu gu cg
a - C u - u
Annotation confidence
High
Do you think this miRNA is real?
Genome context
chr8: 107551244-107551313 [+]
Mature mmu-miR-140-5p
| Accession | MIMAT0000151 |
| Description | Mus musculus mmu-miR-140-5p mature miRNA |
| Sequence | 6 - CAGUGGUUUUACCCUAUGGUAG - 27 |
| Evidence |
experimental
cloned [1-4], Illumina [5-6] |
| Database links |



|
| Predicted targets |


|
Mature mmu-miR-140-3p
| Accession | MIMAT0000152 |
| Description | Mus musculus mmu-miR-140-3p mature miRNA |
| Sequence | 45 - UACCACAGGGUAGAACCACGG - 65 |
| Evidence |
experimental
cloned [2,4], Illumina [5-6] |
| Database links |



|
| Predicted targets |


|
References
|



