Accession
MI0002436
Description
Sus scrofa
ssc-mir-95 precursor miRNA
Literature search
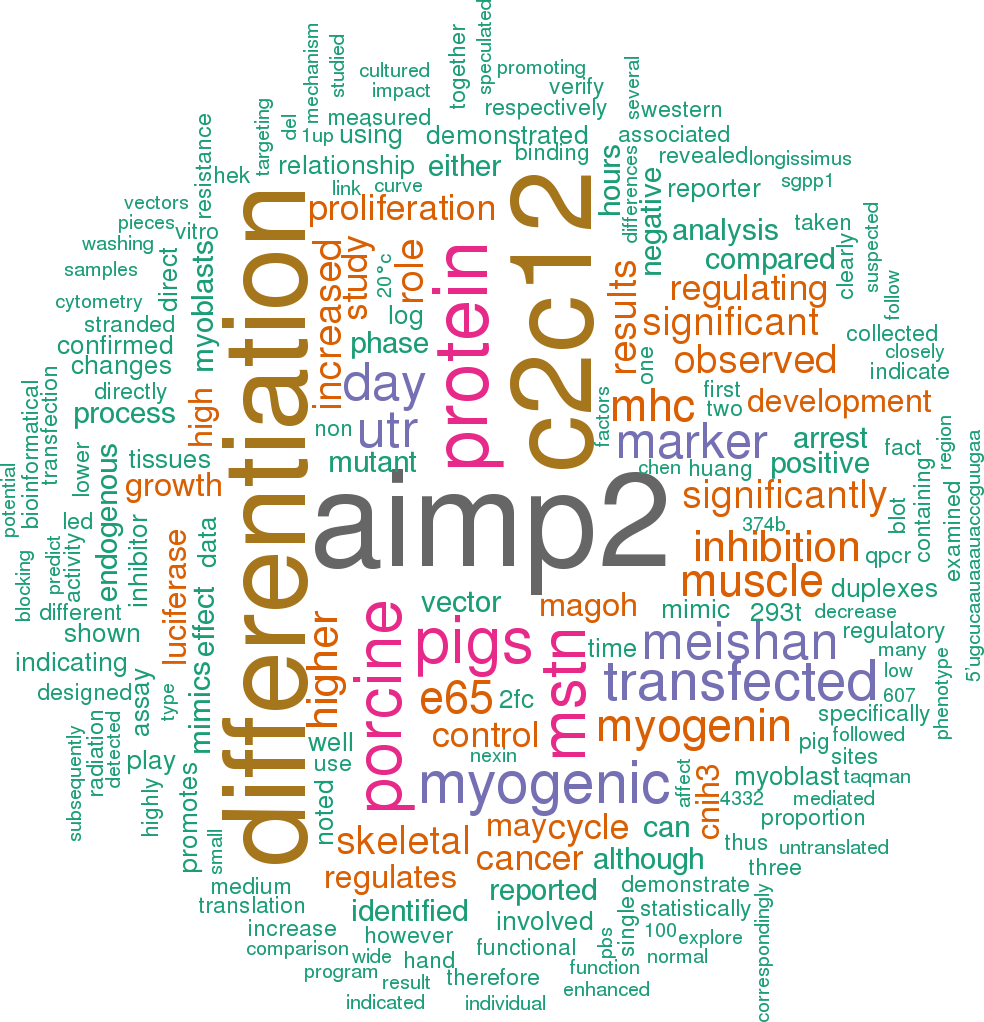
2 open access papers mention ssc-mir-95
(75 sentences)
(75 sentences)
Sequence
33
reads,
22
reads per million, 7 experiments
aacacagugggcgcucaauaaauguuuguugaauugagaugcguuaaaUUCAACGGGUAUUUAUUGAGCAcccacucugug
.(((.((((((.(((((((((((((((((((((((...........))))))))))))))))))))))).)))))).))).
.(((.((((((.(((((((((((((((((((((((...........))))))))))))))))))))))).)))))).))).
Structure
a c c gaga aca aguggg gcucaauaaauguuuguugaauu u ||| |||||| ||||||||||||||||||||||| g ugu ucaccc CGAGUUAUUUAUGGGCAACUUaa c g c A auug
Annotation confidence
Not enough data
Do you think this miRNA is real?
Genome context
chr8: 4277238-4277318 [+]
Mature ssc-miR-95
| Accession | MIMAT0002142 |
| Description | Sus scrofa ssc-miR-95 mature miRNA |
| Sequence | 49 - UUCAACGGGUAUUUAUUGAGCA - 70 |
| Evidence |
experimental
Illumina [2-3] |
References
|


