Accession
MI0004757
Description
Bos taurus
bta-mir-181a-2 precursor miRNA
Literature search
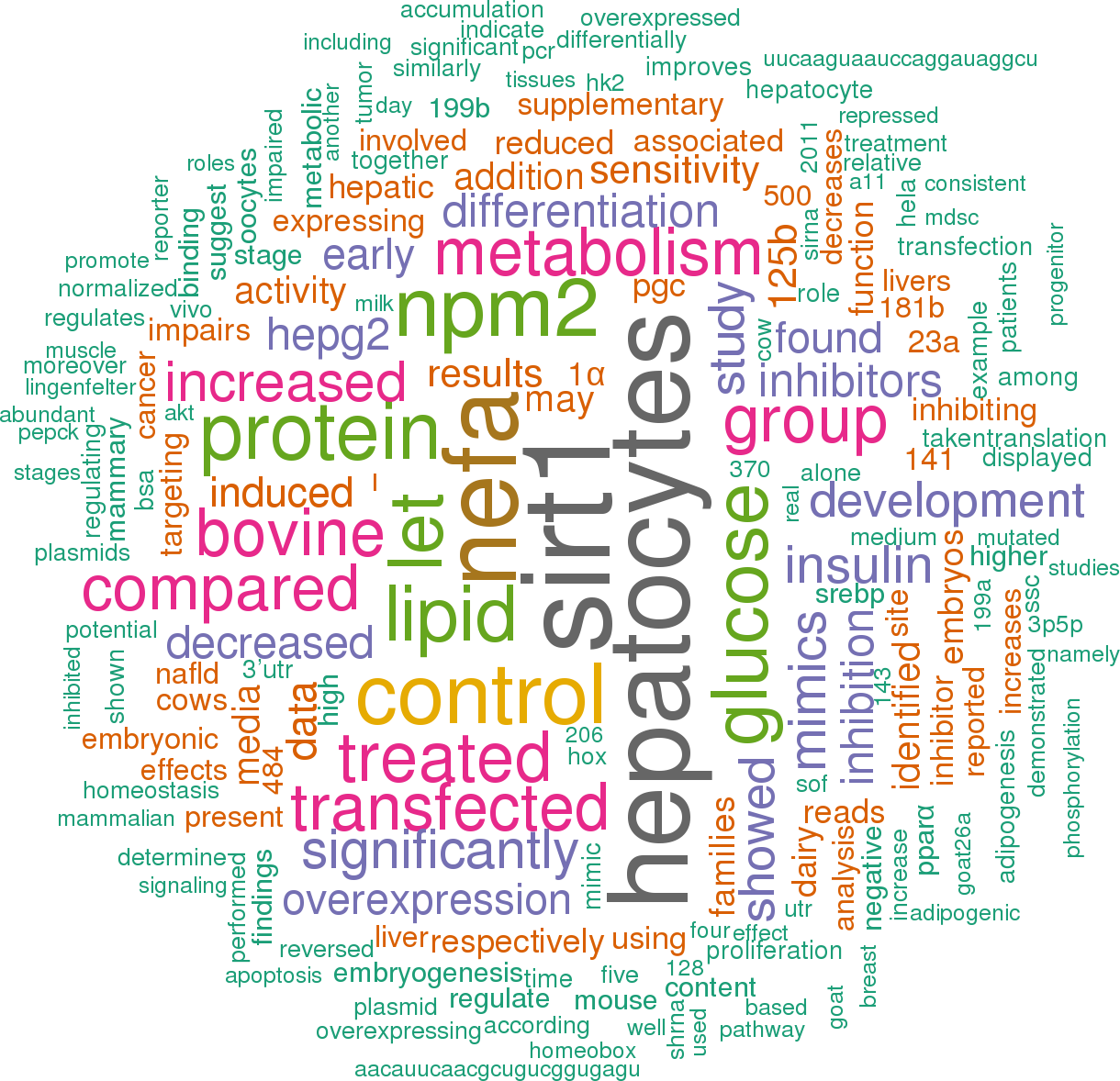
33 open access papers mention bta-mir-181a-2
(245 sentences)
(245 sentences)
Sequence
795630
reads,
8466
reads per million, 78 experiments
ugccagggccaggacccagucuucagaggacuccaaggAACAUUCAACGCUGUCGGUGAGUUugggauuugaaaaaaccaccgaccguugacuguaccuuggguuccuua
.....(((......)))........(((((.(((((((.(((.((((((..(((((((.((((...........))))))))))))))))).))).))))))).))))).
.....(((......)))........(((((.(((((((.(((.((((((..(((((((.((((...........))))))))))))))))).))).))))))).))))).
Structure
ugccagggccaggacccagucuuca c A U CU A ggga
gagga uccaagg ACA UCAACG GUCGGUG GUUu u
||||| ||||||| ||| |||||| ||||||| |||| u
uuccu ggguucc ugu aguugc cagccac caaa u
------------------------a u a c -- - aaag
Annotation confidence
Not enough data
Do you think this miRNA is real?
Genome context
chr11: 95966066-95966175 [+]
Clustered miRNAs
1 other miRNA is < 10 kb from bta-mir-181a-2
| Name | Accession | Chromosome | Start | End | Strand | Confidence |
|---|
Mature bta-miR-181a
| Accession | MIMAT0003543 |
| Description | Bos taurus bta-miR-181a mature miRNA |
| Sequence | 39 - AACAUUCAACGCUGUCGGUGAGUU - 62 |
| Evidence |
experimental
cloned [2] |
References
|



