Accession
MI0004762
Description
Bos taurus
bta-mir-31 precursor miRNA
Literature search
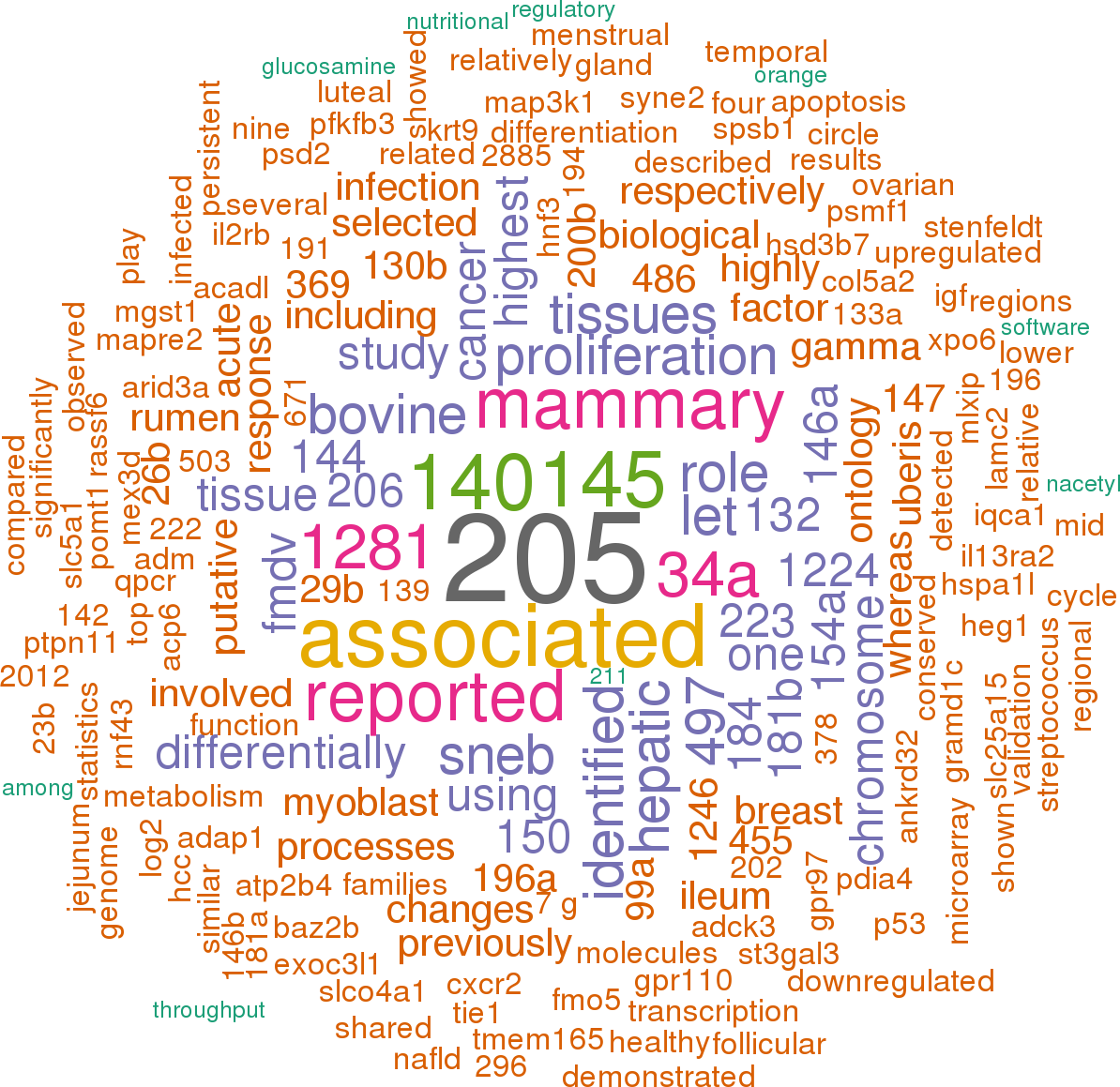
12 open access papers mention bta-mir-31
(44 sentences)
(44 sentences)
Sequence
28539
reads,
271
reads per million, 65 experiments
uccuguaacuuggaacuggagaggAGGCAAGAUGCUGGCAUAGCUguugaacugcgaaccugcuaugccaacauauugccaucucucuuguccg
...........(((.(.(((((((.(((((.(((.(((((((((.(((........)))..))))))))).))).))))).))))))).)))).
...........(((.(.(((((((.(((((.(((.(((((((((.(((........)))..))))))))).))).))))).))))))).)))).
Structure
uccuguaacuu a u A G C -U gaa
gga c ggagagg GGCAA AUG UGGCAUAGC guu c
||| | ||||||| ||||| ||| ||||||||| |||
ccu g ucucucu ccguu uac accguaucg caa u
----------g - u a a a uc gcg
Annotation confidence
High
Do you think this miRNA is real?
Genome context
chr8: 22619494-22619587 [+]
Mature bta-miR-31
| Accession | MIMAT0003548 |
| Description | Bos taurus bta-miR-31 mature miRNA |
| Sequence | 25 - AGGCAAGAUGCUGGCAUAGCU - 45 |
| Evidence |
experimental
cloned [1] |
References
|



