Accession
MI0005051
Description
Bos taurus
bta-let-7g precursor miRNA
Literature search
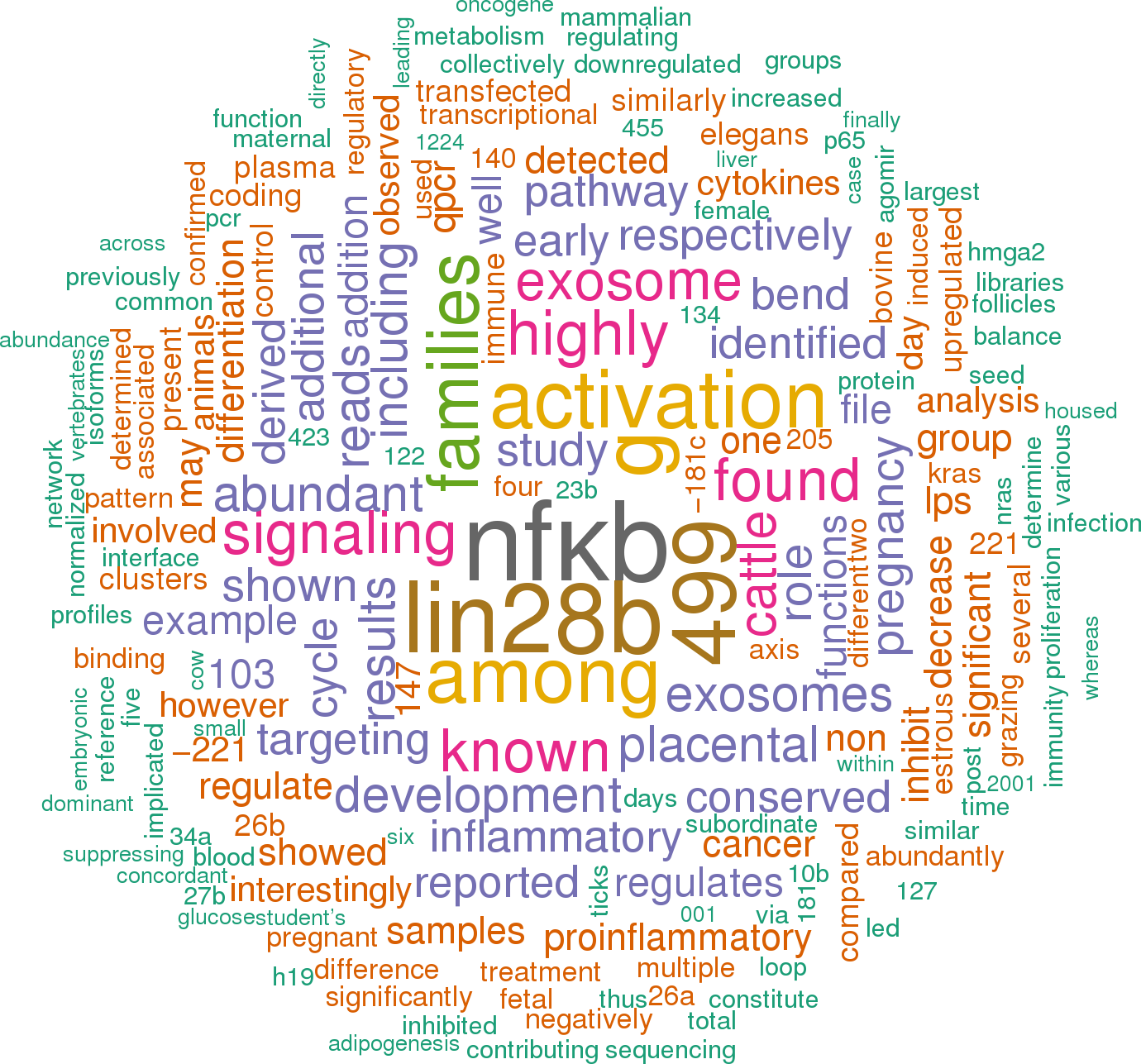
52 open access papers mention bta-let-7g
(205 sentences)
(205 sentences)
Sequence
328959
reads,
1863
reads per million, 76 experiments
aggcUGAGGUAGUAGUUUGUACAGUUugagggucuaugauaccacccgguacaggagauaacuguacaggccacugccuugcc
.(((.((((((((.((((((((((((.....((((.((.((((....))))))..)))))))))))))))).)))))))))))
.(((.((((((((.((((((((((((.....((((.((.((((....))))))..)))))))))))))))).)))))))))))
Structure
a U A ugagg -a a a ggc GAGGUAGU GUUUGUACAGUU gucu ug uacc c ||| |||||||| |||||||||||| |||| || |||| ccg uuccguca cggacaugucaa uaga ac augg c - - c ----- gg - c
Annotation confidence
High
Do you think this miRNA is real?
Genome context
chr22: 49372363-49372445 [+]
Mature bta-let-7g
| Accession | MIMAT0003838 |
| Description | Bos taurus bta-let-7g mature miRNA |
| Sequence | 5 - UGAGGUAGUAGUUUGUACAGUU - 26 |
| Evidence |
experimental
cloned [1-2] |
References
|



