Accession
MI0008220
Description
Sus scrofa
ssc-mir-210 precursor miRNA
Literature search
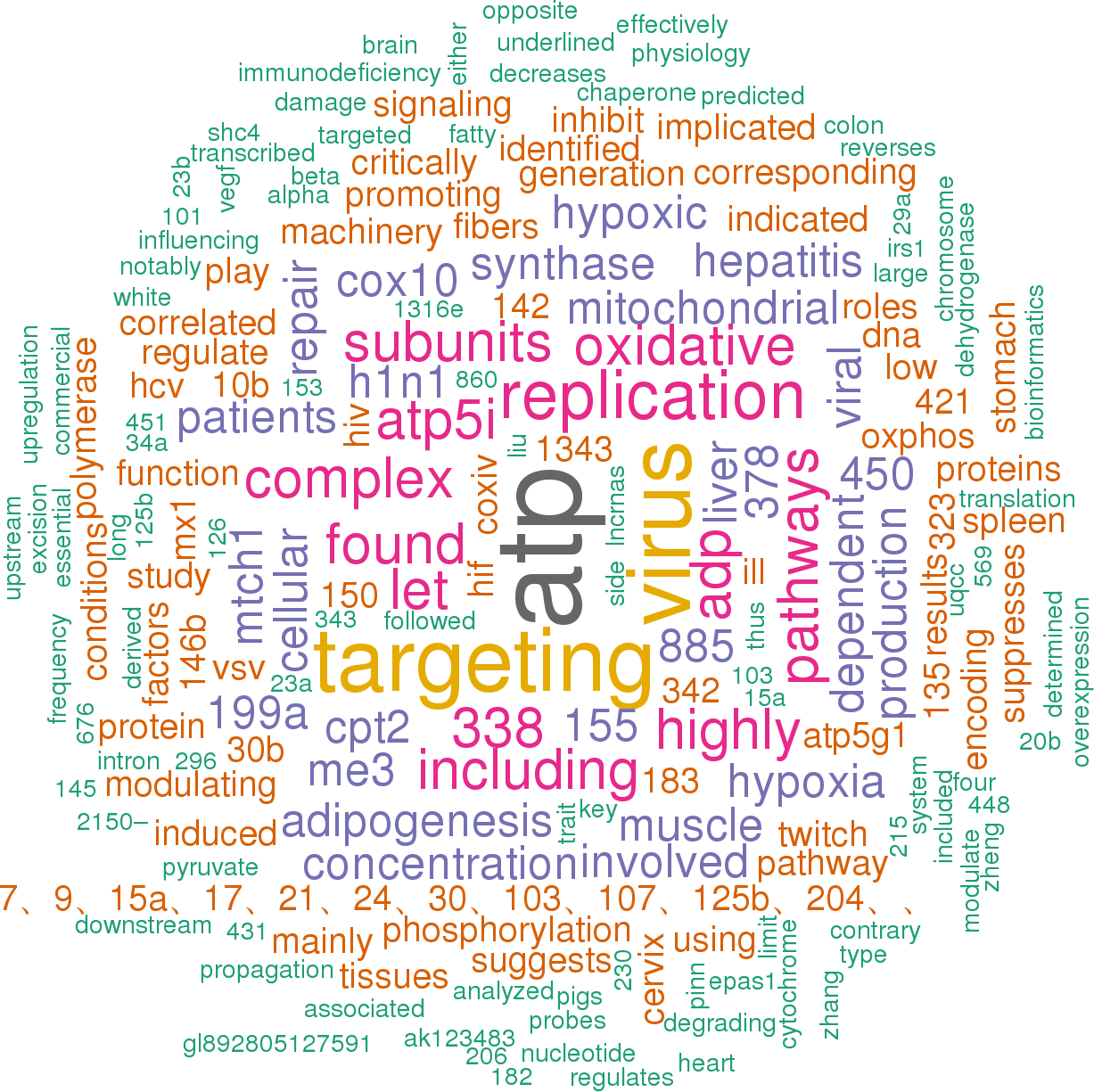
11 open access papers mention ssc-mir-210
(27 sentences)
(27 sentences)
Sequence
14
reads,
7
reads per million, 6 experiments
gcgcagggcagccacugcccaccgcacacugcgcugcuccggacccaCUGUGCGUGUGACAGCGGCUGAucugucc
..((((..(((((.(((..(((((((((.((.((((...))).).)).)))))).))).))).)))))..))))..
..((((..(((((.(((..(((((((((.((.((((...))).).)).)))))).))).))).)))))..))))..
Structure
gc gg a cc - c c - c gcag cagcc cug cac cgcaca ug g cug |||| ||||| ||| ||| |||||| || | ||| u uguc GUCGG GAC GUG GCGUGU ac c ggc cc uA C -A U C c a c
Annotation confidence
Not enough data
Do you think this miRNA is real?
Genome context
chr2: 339074-339149 [+]
Mature ssc-miR-210
| Accession | MIMAT0007761 |
| Description | Sus scrofa ssc-miR-210 mature miRNA |
| Sequence | 48 - CUGUGCGUGUGACAGCGGCUGA - 69 |
| Evidence |
experimental
cloned [1], Illumina [2-4] |
References
|


