Accession
MI0013107
Description
Sus scrofa
ssc-mir-181b-1 precursor miRNA
Literature search
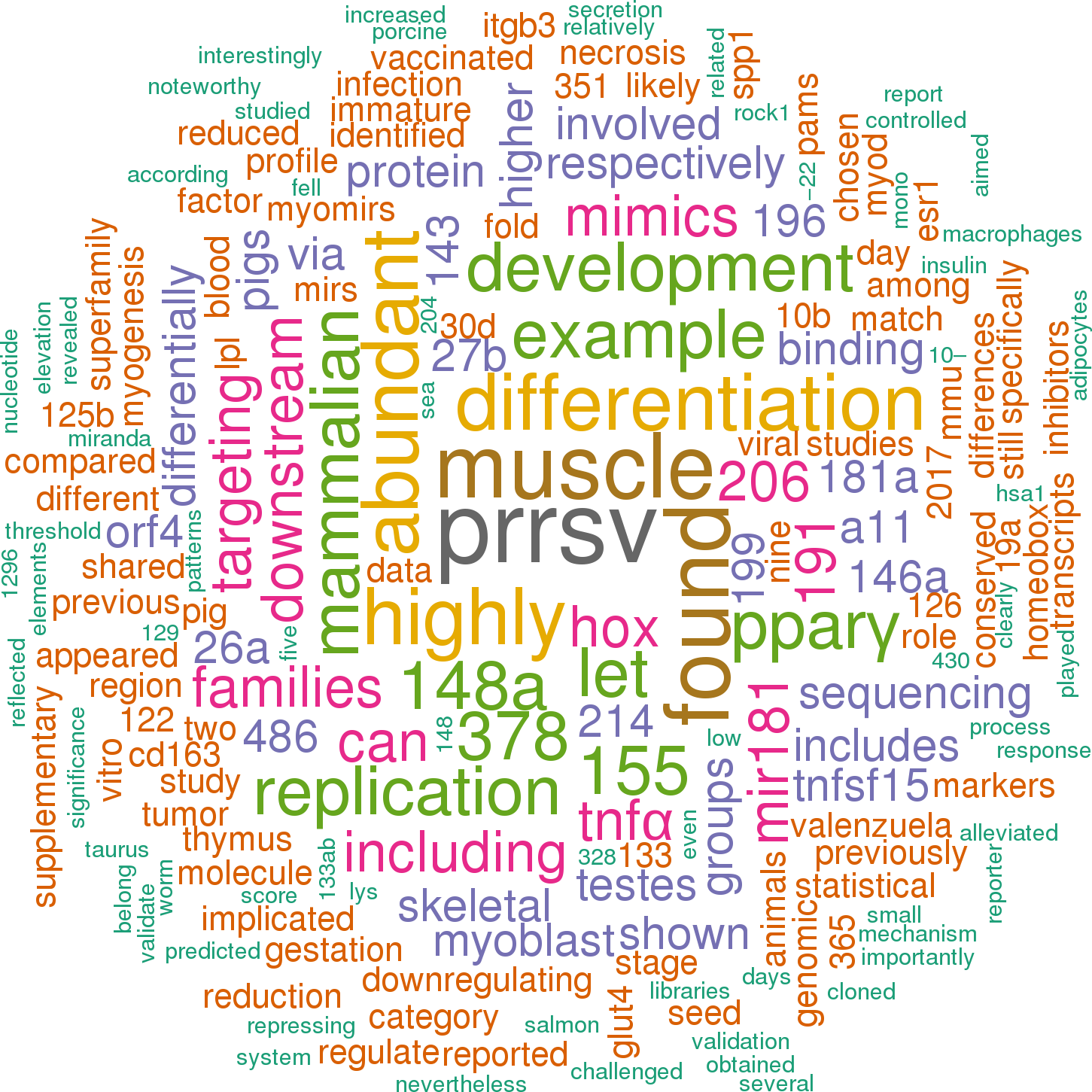
25 open access papers mention ssc-mir-181b-1
(47 sentences)
(47 sentences)
Sequence
60
reads,
16
reads per million, 11 experiments
ggucacaaucAACAUUCAUUGCUGUCGGUGGGUUgaacuguguagacaagcucacugaacaaugaaugcaacuguggccc
(((((((.....(((((((((...((((((((((....(((....))))))))))))).)))))))))....))))))).
(((((((.....(((((((((...((((((((((....(((....))))))))))))).)))))))))....))))))).
Structure
- aucAA CUG gaac g ggucaca CAUUCAUUG UCGGUGGGUU ugu u ||||||| ||||||||| |||||||||| ||| ccggugu guaaguaac agucacucga aca a c -caac --a ---- g
Annotation confidence
Not enough data
Do you think this miRNA is real?
Genome context
chr10: 26426857-26426936 [-]
Clustered miRNAs
1 other miRNA is < 10 kb from ssc-mir-181b-1
| Name | Accession | Chromosome | Start | End | Strand | Confidence |
|---|
Mature ssc-miR-181b
| Accession | MIMAT0002126 |
| Description | Sus scrofa ssc-miR-181b mature miRNA |
| Sequence | 11 - AACAUUCAUUGCUGUCGGUGGGUU - 34 |
| Evidence |
experimental
Illumina [1-3] |
References
|


