Accession
MI0013172
Description
Sus scrofa
ssc-mir-125b-1 precursor miRNA
Literature search
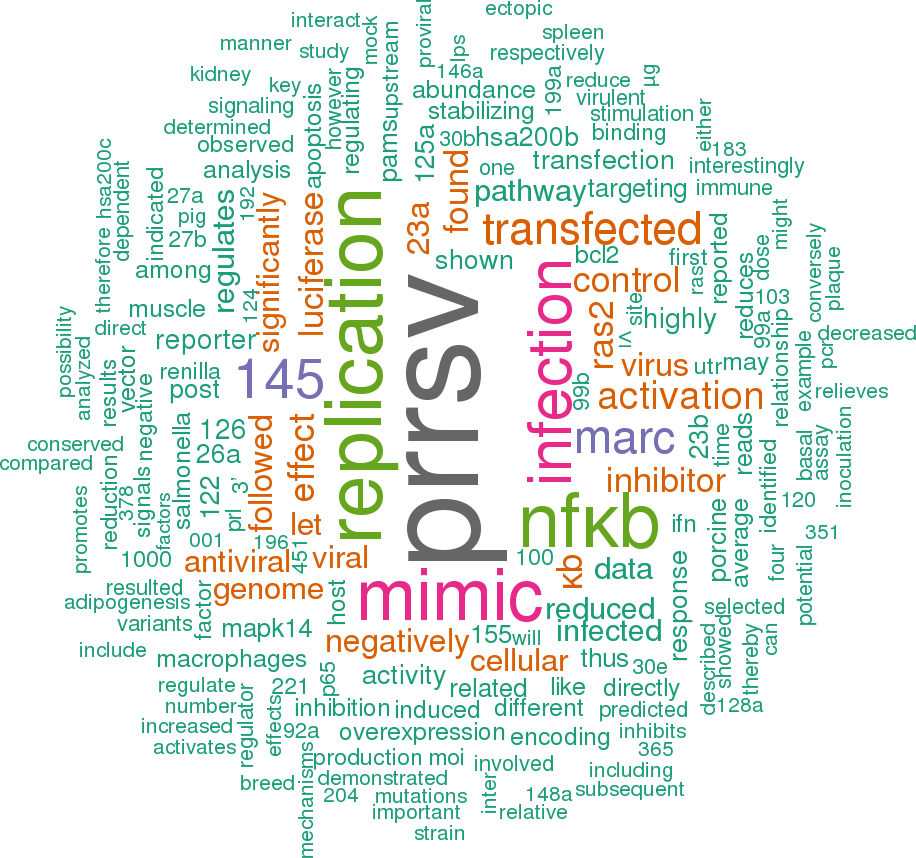
23 open access papers mention ssc-mir-125b-1
(162 sentences)
(162 sentences)
Sequence
1817
reads,
406
reads per million, 13 experiments
ccucucagUCCCUGAGACCCUAACUUGUGAuguuuaccguuuaaauccacggguuaggcucuugggagcugcgagccgug
.(((.((((.(((((((.(((((((((((..(((((.....))))).))))))))))).))))))).)))).))).....
.(((.((((.(((((((.(((((((((((..(((((.....))))).))))))))))).))))))).)))).))).....
Structure
----c u C C Au c
cuc cagU CCUGAGA CCUAACUUGUG guuua c
||| |||| ||||||| ||||||||||| ||||| g
gag gucg ggguucu ggauugggcac uaaau u
gugcc c a c -c u
Annotation confidence
Not enough data
Do you think this miRNA is real?
Genome context
chr9: 54334908-54334987 [-]
Mature ssc-miR-125b
| Accession | MIMAT0002120 |
| Description | Sus scrofa ssc-miR-125b mature miRNA |
| Sequence | 9 - UCCCUGAGACCCUAACUUGUGA - 30 |
| Evidence |
experimental
Illumina [1-3] |
References
|


