Accession
MI0009851
Description
Bos taurus
bta-mir-500 precursor miRNA
Literature search
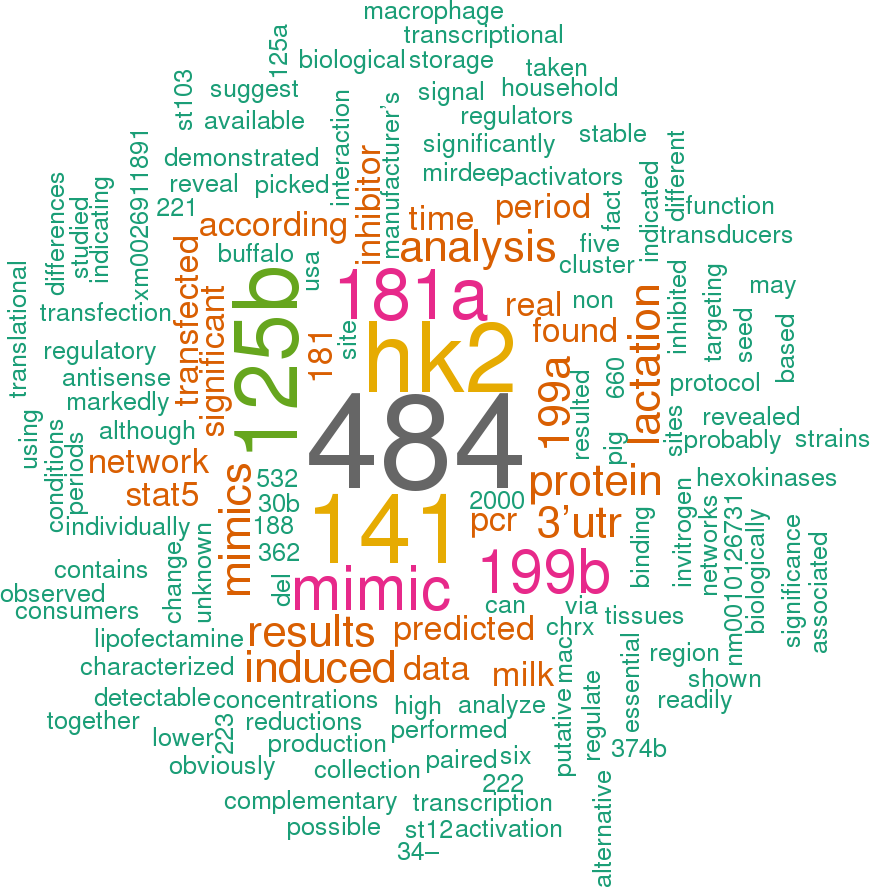
4 open access papers mention bta-mir-500
(16 sentences)
(16 sentences)
Sequence
9255
reads,
190
reads per million, 76 experiments
gcucccccucucUAAUCCUUGCUACCUGGGUGAGAgugcuuucugaaugcaaugcaccugggcaaggauucugagagagggagc
((((((.(((((.(((((((((...(((((((....(((.........)))...))))))))))))))))..))))).))))))
((((((.(((((.(((((((((...(((((((....(((.........)))...))))))))))))))))..))))).))))))
Structure
c -U UAC AGAg uuu
gcuccc cucuc AAUCCUUGC CUGGGUG ugc c
|||||| ||||| ||||||||| ||||||| ||| u
cgaggg gagag uuaggaacg gguccac acg g
a uc --- -gua uaa
Annotation confidence
High
Do you think this miRNA is real?
Genome context
chrX: 93948610-93948693 [+]
Clustered miRNAs
5 other miRNAs are < 10 kb from bta-mir-500
| Name | Accession | Chromosome | Start | End | Strand | Confidence |
|---|
Mature bta-miR-500
| Accession | MIMAT0009337 |
| Description | Bos taurus bta-miR-500 mature miRNA |
| Sequence | 13 - UAAUCCUUGCUACCUGGGUGAGA - 35 |
| Evidence |
experimental
cloned [3-4] |
References
|


